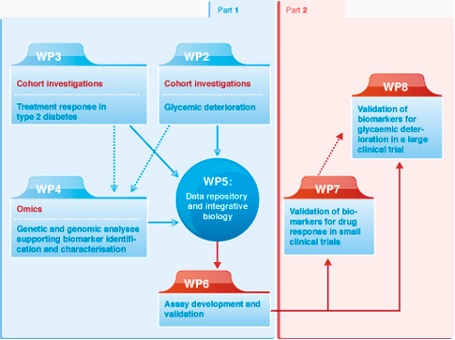
Studies of WP3 focus on therapeutic interventions to identify biomarkers that predict response or non-response to metformin, sulphonylureas, GLP-1R agonists, and early remission from obesity surgery. We utilised the large cohorts available to model drug response in our populations to complete the phenotyping of these already well-characterised subjects using standard protocols.
Some patients with type 2 diabetes do not respond to medication and others show an extreme response, or extreme adverse reaction. If biomarkers can be identified that predict response, these could be used to target therapy by patient stratification. This may allow, for example, earlier intervention with drugs currently reserved for second or third-line treatment in those predicted not to respond to traditional first line (metformin) or second-line (sulphonylurea) therapy.
The major objective of this work package is to develop treatment response cohorts for provision to the genomics work package (WP4), and the integrative biology work package (WP5) with a view to identifying biomarkers that predict extreme treatment response (or side effect) to a number of therapeutic interventions.
This approach should yield novel biomarkers that will help target current and future medical therapies focused on preventing and treating type 2 diabetes.